OIL PALM
The oil palm (Elaeis guineensis Jacq.) is a tropical perennial crop originating in Central and Western Africa in the humid lowland tropics, within ±10˚ latitude of the equator. Oil palm is a monocotyledonous tree belonging to the Family Arecacea, tribe Cocoseae, and subtribe Elaeidinae. This subtribe consists of the genus Elaeis. The genus name, Elaeis, is from the Greek ‘elaia’ for olive. The African oil palm’s specific name, guineensis, refers to the tree’s discovery by Jacquin in the Gulf of Guinea area. The genus also contains E. oleifera HBK (the South American oil palm). The major interest in oil palm is the high oil content of the mesocarp and kernel of the fruits that are produced in heavy bunches. The plant is monoecious with out-crossing maintained through successive cycles of male and female inflorescence production on a single palm. The length of the male and female cycles varies widely according to genotype and environment. At maturity, the fruit is red-brown and consists of the pulp, the shell, and the kernel. The pulp yields edible, orange-red oil commonly known as palm oil. The endosperm or kernel when crushed produces clear yellowish oil that is known as palm kernel oil and is similar to coconut oil. Palm oil and its refined derivatives, palm olein and palm stearin, are the major commercial products of oil palm.
The plant generation time is long, with seeds taking around 100–120 days to germinate, including a heat-treatment, followed by 10–12 months in the nursery before the young seedlings are ready for field planting. The oil palm starts to bear fruit after 2–3 years of field planting and approaches maturity at around 10 years. The economic life of plantings varies from 20–30 years, depending on local conditions. Of all oil-bearing plants, the oil palm produces the most oil per hectare per year.
Biotechnology of Oil Palm
The potential of oil palm biotechnology has been reviewed a number of times and one area where considerable progress has been made is clonal propagation. As oil palm has only a single vegetative meristem, it is not possible to replicate elite genotypes by cuttings or graftings. The absence of fully inbred lines means that it is also not possible to commercially propagate elite individual oil palm genotypes by seed. These two constraints led to the development of a clonal propagation system based on somatic embryogenesis. The development of suspension culture systems for the production of somatic embryos and of cryopreservation techniques for their storage also offers the possibility of mass production of artificial seed.
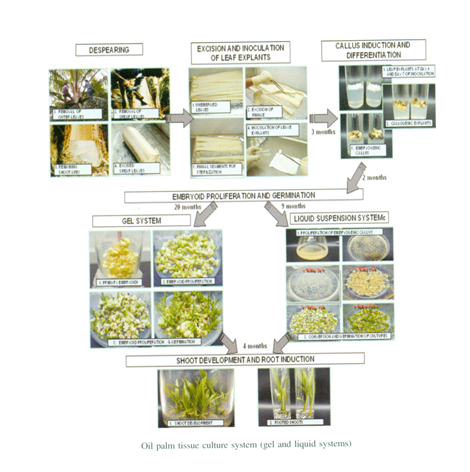 |
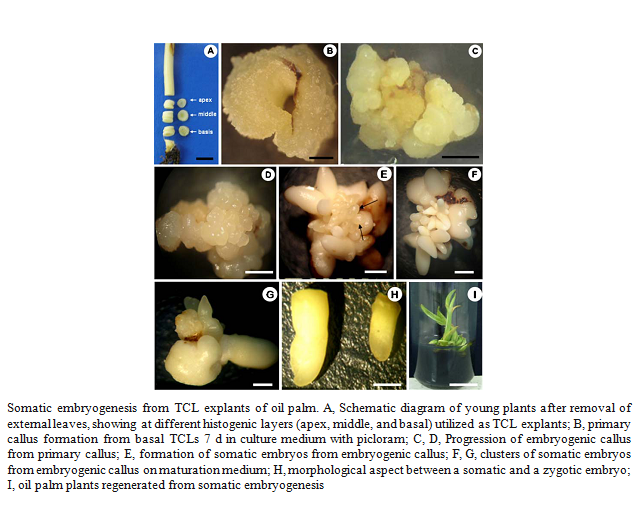
An efficient procedure has been developed for inducing somatic embryogenesis and regeneration of plants from tissue cultures of oil palm. Thin transverse sections (thin cell layer explants) of different position in the shoot apex and leaf sheath of oil palm were cultivated in Murashige and Skoog (MS) medium supplemented with 0–450 μM picloram and 2,4-D with 3.0% sucrose, 500 mg L−1 glutamine, and 0.3 gL−1 activated charcoal and gelled with 2.5 gL−1 Phytagel. Embryogenic calluses were maintained on a maturation medium composed of basal media, plus 0.6 μM NAA and 12.30 μM 2iP, 0.3 gL−1 activated charcoal, and 500 mg L−1 glutamine, with subcultures at 4-week intervals. Somatic embryos recovered were converted to plants on MS medium with macro- and micro-nutrients at half-strength, 2% sucrose, and 1.0 gL−1 activated charcoal and gelled with 2.5 gL−1 Phytagel.
 |
In addition to enabling the production of large numbers of elite genotypes, tissue culture methods provide the basis for research on genetic transformation. The first report of transient expression in oil palm tissues, using the biolistics approach, was presented at the International Oil Palm Congress (PIPOC) in 1993 and significant progress has been made since. In many species, transformation approaches based on Agrobacterium tumefaciens have been developed. Compared to the biolistics approaches, this method generates simpler and lower copy inserts in transformed plants. This is important for stability of transgene expression. Agrobacterium-mediated transformation also produces unlinked inserts in a reasonable number of transformants, potentially facilitating the segregation of the selective marker away from the gene of interest.
 |
Transgenic plants
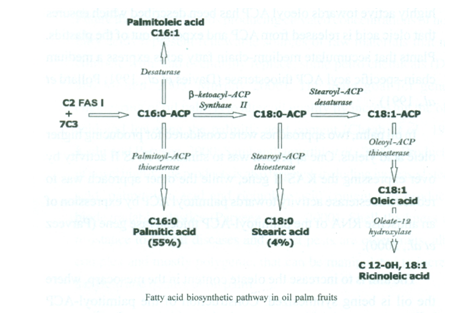
The main goal for genetic engineering of oil palm is to alter oil quality by increasing the oleic acid and lowering the palmitic acid content. Production of novel high-value products is also targeted, including increased stearic acid, palmitoleic acid and ricinoleic acid content and producing biodegradable plastics. Shell thickness and resistance to fungal diseases and insect pests are other traits, albeit complex and mostly polygenic, that can be manipulated to increase the yield and quality of oil palm.
From induction of callus to production of field-planted palms producing a yield can take 8-10 years. Transgenic approaches would raise potential containment issues of the modified gene (as well as issues of safeguarding the research investment) and in some countries theft of material from nurseries is common. The targeting of transgenes to the plastid might resolve the issue of GM pollen release and might also improve the efficiency with which expression takes place. However, the potential to produce a high-oleate palm oil and other novel oils for nutraceutical or industrial markets and the possibility of tackling some currently intractable problems makes transgenic technology inviting.
 |
Genetic Fingerprinting
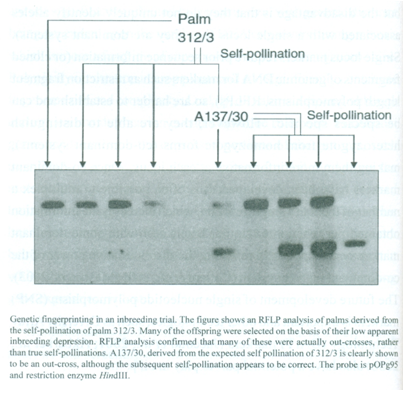
There are very limited numbers of simple morphological markers present in commercial breeding material. Initial genetic fingerprinting was carried out using isozymes and RFLP markers. Genetic fingerprinting has been used successfully in a number of programs, as in the Unilever clonal propagation program at Unifield, Bedford, UK. Another example of the value of fingerprinting is provided from the Unilever inbreeding trial planted at Binga, Democratic Republic of Congo, in 1973. The palm Bg. 312/3 was self-pollinated to generate a population with an inbreeding coefficient of Fx = 0.56, reflecting the chance of two loci being identical by descent. The progeny and subsequent crosses were planted in numerous countries as part of a combined breeding program. Palms within this particular progeny (Bg. 143) were further selected due to their low levels of apparent inbreeding depression. Genetic fingerprints confirmed that a majority of the palms tested were actually out-crosses, including the parent of the first mapping cross.
Due to the development of significant numbers of publicly available microsatellite, simple sequence repeat (SSRs) markers it is now possible to apply genetic fingerprinting at key points in both breeding and commercial palm production to monitor systems and to ensure quality control. SSRs are probably the best available tool currently for routine fingerprinting via multiplexed pools, making the approach cheap and highly discriminative.
 |
Diversity Analysis
Diversity analysis is a potentially powerful approach, particularly in species where early breeding records are missing or unclear and in species which are natural out-breeders, where controlled crossing is not always completely effective. While molecular markers do not remove the need to field-evaluate the traits of particular accessions, they do offer an additional level of information which potentially allows the minimum number of desirable accession palms to be introduced to capture the maximum amount of genetic diversity. A number of diversity analyses have been reported in the literature, including those based on (1) isozyme markers, (2) RFLP markers, (3) amplified fragment length polymorphisms (AFLP) plus RFLP markers, (4) randomly amplified polymorphic DNA (RAPD) markers, (5) AFLP markers, and (6) AFLP plus isozyme markers. The development of retrotransposon-based marker systems, both generic and an oil palm specific copia-based system have significant potential for phylogenetic reconstruction, with retrotranposon movement being an essentially irreversible process.
Linkage Mapping
The development of markers close to the shell-thickness gene, preferably flanking markers, would allow shell thickness to be predicted before field planting. This would have relevance for breeding trials, allowing potentially female sterile pisifera palms to be planted separately from other shell-types (as they can exhibit excessive vegetative vigor) or even allowing a robust quality control test of tenera seed lots for commercial production. The recent publication of a comprehensive genetic map combining SSR and AFLP markers represents a significant step forward for oil palm molecular genetics. A number of the oligogenic traits in oil palm have also been tentatively placed onto the genetic linkage maps in addition to shell-thickness, including virescens and a component of crown disease. The existence of a good genetic map is an important prerequisite for the development of a physical map.
QTL Analysis
The majority of traits of commercial importance are thought to be polygenic and often have significant environmental interactions. Estimating heritabilities in well designed experiments can assist in understanding how significant the genotype (G) effect is compared with the environmental (E) influence. However, even the use of bunch analysis to simplify the individual components of oil yield has still not produced clearly identifiable major gene effects beyond those already identified. This may be because genetic analysis is complicated by significant G x E interactions masking either a relatively simple mode of inheritance, pleiotropic effects, genuinely polygenic inheritance, epistasis, dominance, or disequilibrium effects. One approach to try to resolve this problem is through quantitative trait locus (QTL) analysis.
The major disadvantage of QTL analysis is that it detects only QTL that are segregating in a particular cross. Whether the genetic markers are fully informative also has an effect on the completeness of the genetic data for comparison to trait data. However, the advantages are also significant. When the QTL analysis is carried out in material of direct commercial or breeding interest, there is the potential to select for the presence of a QTL using flanking DNA markers in subsequent breeding.
 |
Developing BAC Libraries for Oil Palm
The ability to maintain large fragments of a plant genome artificially is one of the key steps to developing genomic resources for a species. One system is the cosmid system which has been used for oil palm, another is the bacterial artificial chromosome (BAC) system. BAC vectors are capable of cloning and stably maintaining DNA fragments of up to 300 kb in Escherichia coli, exhibit low levels of chimerism, and are amenable to high-throughput and economical DNA purification, and thus, large-scale genome sequencing. For oil palm, with a haploid genome size of 980 Mb, an average BAC insert size of 120 kb would require 8,000 bacterial clones to represent one haploid equivalent of the genome. Having this basic resource is critical for genomic approaches. The first BAC library reported for oil palm was constructed using HindIII endonuclease; nine tropical crop libraries were constructed in the BACTROP program. The potential for using methylation-sensitive restriction endonucleases to construct BACs targeting the lower-copy and coding regions has also been investigated with some success. Having an arrayed and searchable BAC resource for oil palm is particularly important for map-based cloning approaches. One major concern with such approaches is the lack of consistent relationship between genetic distance (cM) and physical distance (Mb) along chromosomes. In many species, centromeric regions of the chromosome are highly suppressed for recombination, which could make cloning genes in these regions by map-based cloning difficult, if not impossible. Initial map positions for shell-thickness, however, suggest that it is distal on the chromosome, making such an attempt possible.
Physical Mapping and Genome Sequencing
Eventually, the ideal situation would be to have the entire genome of oil palm sequenced. One staging post to doing this in an ordered and coherent fashion is the development of a physical map. This essentially involves combining genetic mapping data with the position of physical clones, such as BACs. Once this has been done, it is possible to choose and sequence individual clones in the most efficient way to produce a contiguous genome sequence. BAC or cosmid contigs can be generated using a variety of approaches and linked into the genetic map. A comprehensive program has just been completed to create methylation filtration sequence data for oil palm. The methodology involves creation of a representative genomic library in a bacterial strain which does not tolerate methylation of DNA bases. This leads to propagation of clones that come from non-methylated regions of the genome, which are expected to be the low-copy and coding regions. The only real disadvantage of this technique is that methylation is so common in plant species that the clone sizes, i.e., the continuous sequence obtained, is limited, usually averaging in the 1–2 kb range. Methyl filtration sequence data represents an important resource for oil palm genomics and should assist the development of physical maps of the coding regions. Such sequence and clones can be used as one form of “marker” which links together the genetic and physical maps, as well as providing detailed sequence information for the species of interest. Clearly, a complete physical map would also enhance map-based walking approaches, by allowing a series of markers to be developed and tested across the region of interest, without the step-by-step BAC walking. The development of a detailed genetic map, ordered BAC libraries, EST resources, and methylation filtration sequence information for oil palm makes the development of a physical map for the oil palm genome now feasible.
Genome Structure and Conserved Synteny
Another approach which can also assist in this process, while generating further information on how the genome of oil palm is organized, is molecular cytogenetics. Work in Heslop-Harrison’s lab in Leicester University (UK) has pioneered attempts to visualize the oil palm genome using genomic in situ hybridization (GISH) and fluorescent in situ hybridization (FISH) techniques. A number of repeat sequences used as probes have permitted the chromosomes of oil palm to be distinguished and a comparison of repeat sequence distribution made. This has been extended to examine oil palm chromosomes during meiosis. Such examination would be a particularly useful selection tool for introgression of desirable E. oleifera genes into advanced E. guineensis germplasm. Retrotransposons have already been shown to be important components in the genome of oil palm with evidence for divergent evolution of different classes of these sequences since the split between E. guineensis and E. oleifera.
Functional Genomics
Functional genomics in oil palm has focused on developing and analyzing EST collections and using this information to explore the potential for using microarrays for transcript analysis.
Expressed Sequence Tag (EST) Development
A recent MPOB-MIT collaborative program has developed the first significant numbers of ESTs for oil palm. Six thousand five hundred are currently available on the MPOB site in a searchable database, a further 2,411 have been reported by CIRAD-CP, and about 20,000 have been generated from an oil palm tissue culture cDNA library. These have been used to generate a microarray chip containing 3,806 oil palm gene clones. This is an important step on the way to comprehensive transcriptomics resources.
Transcriptomics
The original technology used to examine patterns of gene expression in a tissue was northern analysis, where radiolabelled probes were hybridized to total RNA, or mRNA, which had been size-separated and immobilized onto a filter. A few dozen samples could be interrogated with a few genes at a time. Development of a 3,806 clone array spotted onto a slide allows a thousand times more genes to be evaluated by labeling the mRNA itself, having an immobilized gene probe. Initially, this was used to compare normal and abnormal tissue culture material, but it has important potential for other applications.
Bioinformatics
Bioinformatics is critical for the successful application of genomics and functional genomics technologies. This is literally true in the sense that it is impossible to analyze the amounts of data generated in a coherent fashion without dedicated tools and in the more general sense that cross species comparisons offer an extremely powerful approach for working in crops where resources and information are more limited. It would be true to say that the tools immediately needed for oil palm bioinformatics tools already exist in other crop and model species and could be easily applied to oil palm – the limitation in oil palm is currently the amount of sequence-based data, not the tools to handle it.
Source: Dr.V.Ponnuswami, PhD, PDF (Taiwan), Former Dean & Professor (Horticulture), Horticultural College & Research Institute, Tamil Nadu Agricultural University, Coimbatore
|
.png)
.png)